Beetle artwork by Meg McConnell
"Nothing in biology makes sense except in the light of evolution" - Dobzhansky 1973
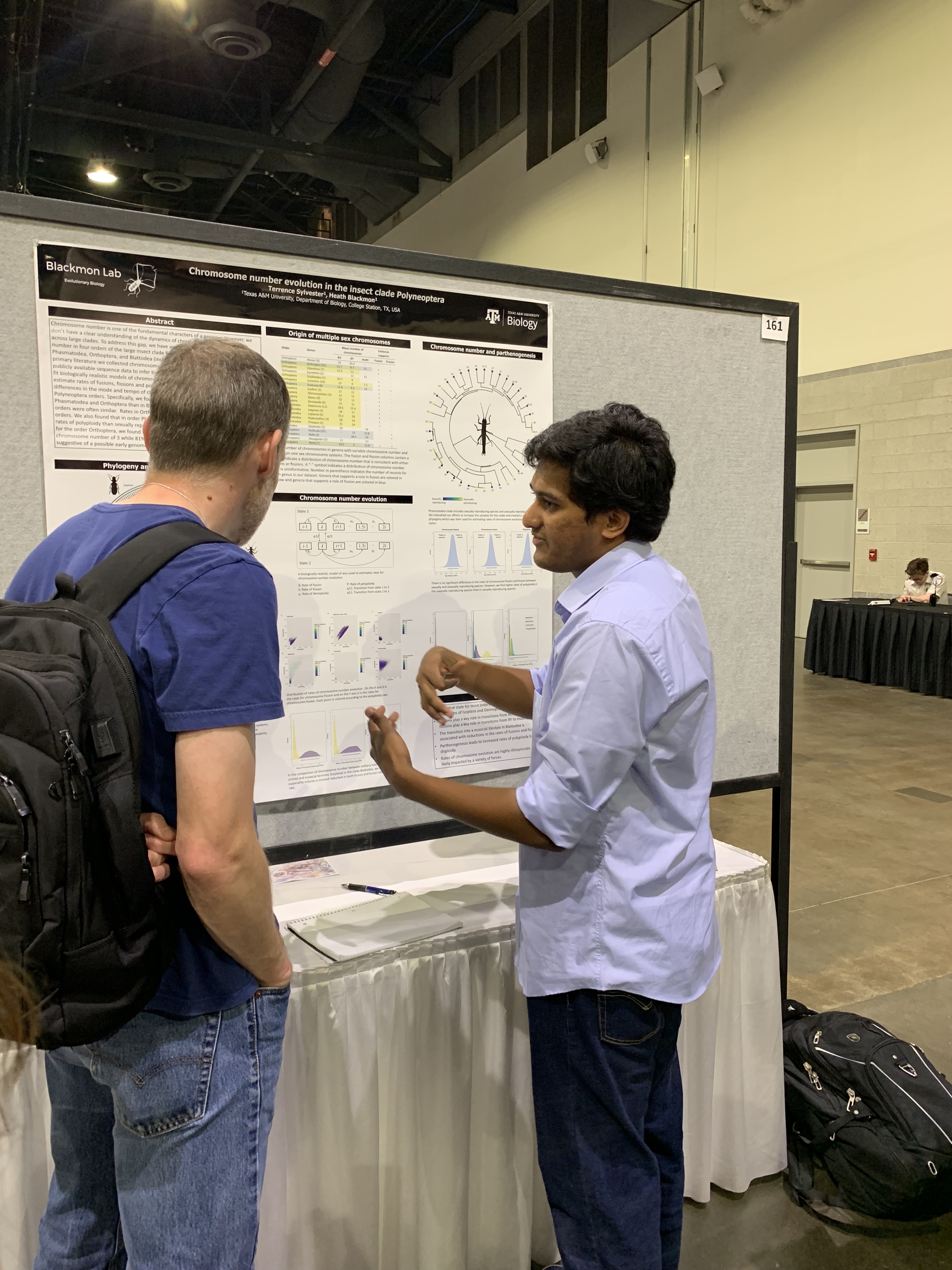
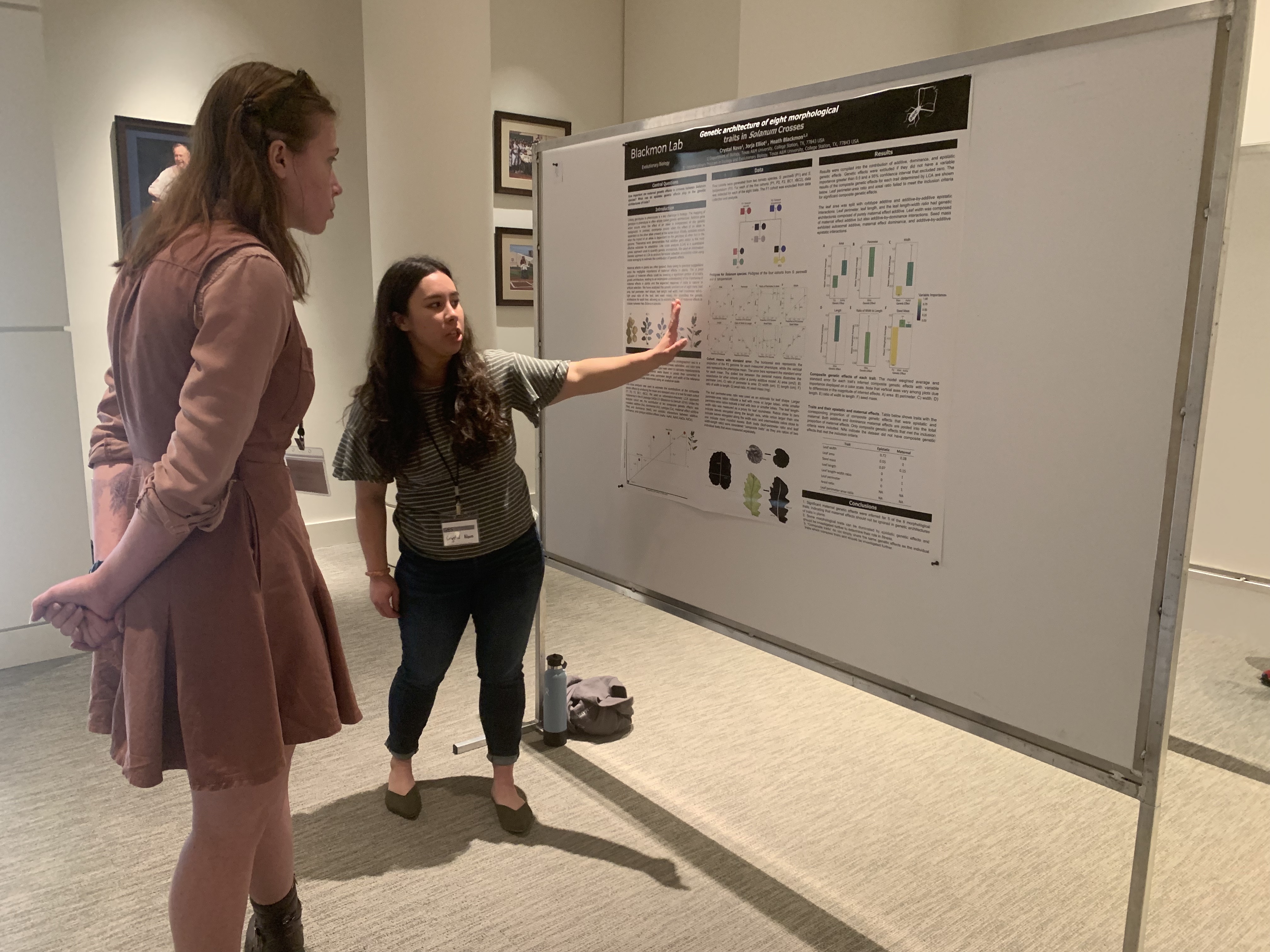
Research
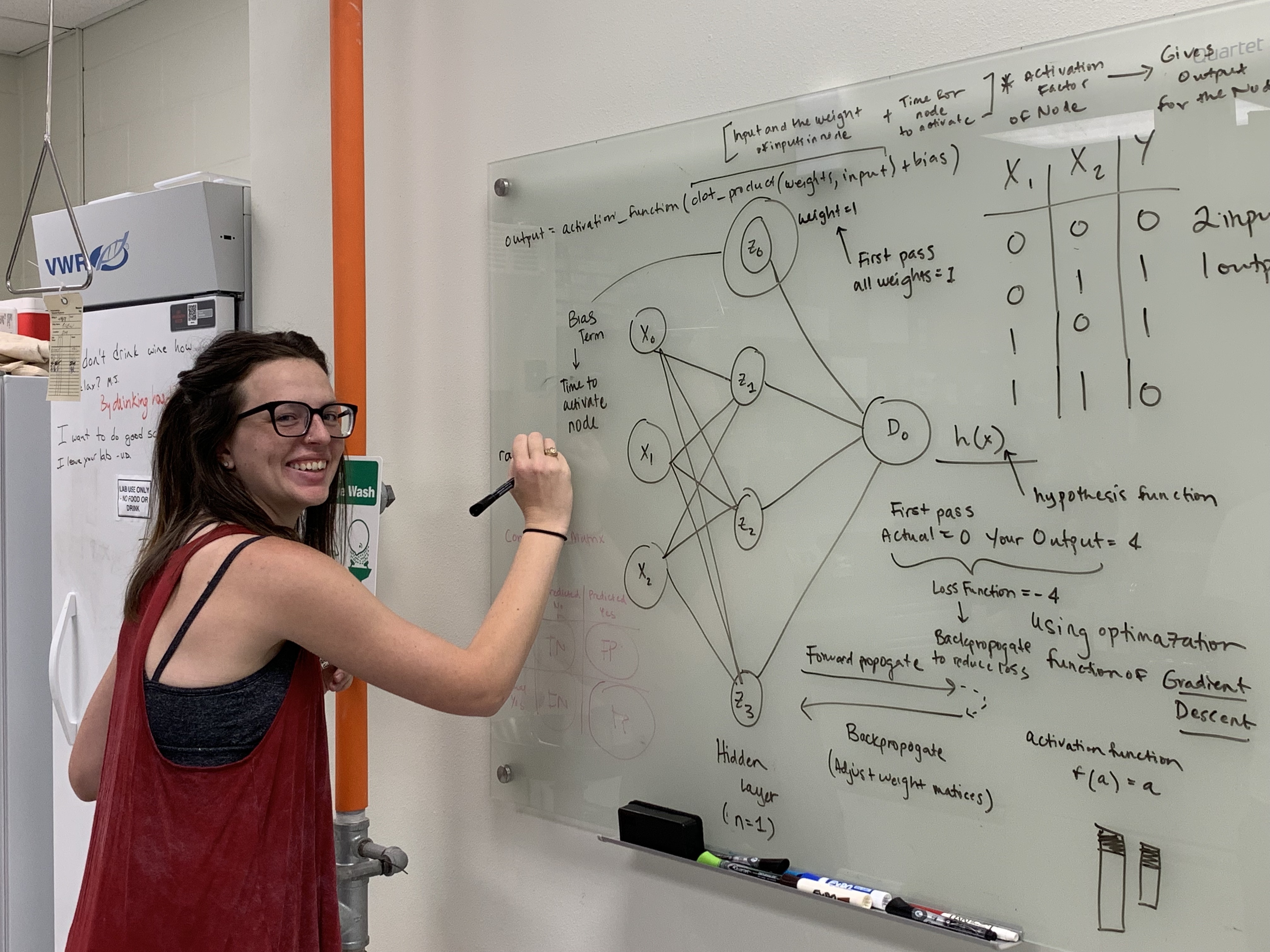
Genome evolution, sex chromosomes, and computational biology
Publications
Papers from 2012 to present, live from ORCID
Team
PI, grad students, staff, and alumni
Resources
R packages, databases, teaching materials
Join Us
Expectations, calendar, and how to apply
Lab Life
Photos, fieldwork, and fun
Study Systems

Beetles
Karyotype evolution and sex chromosome systems across Coleoptera — the most species-rich order of animals.

Tomatoes
Chromosome number variation and its link to adaptation in Solanaceae, including wild and domesticated species.

Betta Fish
Genetics of aggression, color, and sexual selection — and how domestication shapes phenotypic evolution.

Chickens
Sex-linked traits and sexual antagonism in a model system for understanding domestication.

Crabs
The evolution of freshwater invasion and its genomic consequences across Brachyura.

Mammals
Broad-scale comparative analyses of sex chromosome evolution and karyotype diversity.